Genomic Standards Consortium
The Genomic Standards Consortium (GSC) is an open-membership working body formed in September 2005. The aim of the GSC is making genomic data discoverable. The GSC enables genomic data integration, discovery and comparison through international community-driven standards.
This project is maintained by GenomicsStandardsConsortium
MIxS Extensions (Packages)
The MIxS standard is designed as an overarching framework, in order to create a single entry point to all minimum information checklists and extensions from the GSC. MIxS includes the technology-specific checklists from the previous MIGS, MIMS and MIMARKS standards, provides a way of introducing additional checklists such as MISAG, MIMAG and MIUViG, and also allows annotation of sample data using extensions.Initially introduced as Environmental extensions to standardize sets of measurements and observations describing particular habitats that are applicable across all GSC checklists, we now refer to them as just extensions to allow for the future expansion into non-environmental extensions such as the ancient-DNA specific extension current under review. With this modular and extensible framework, both horizontal (i.e new checklists) and vertical (i.e. new extensions) expansion is possible and encouraged.
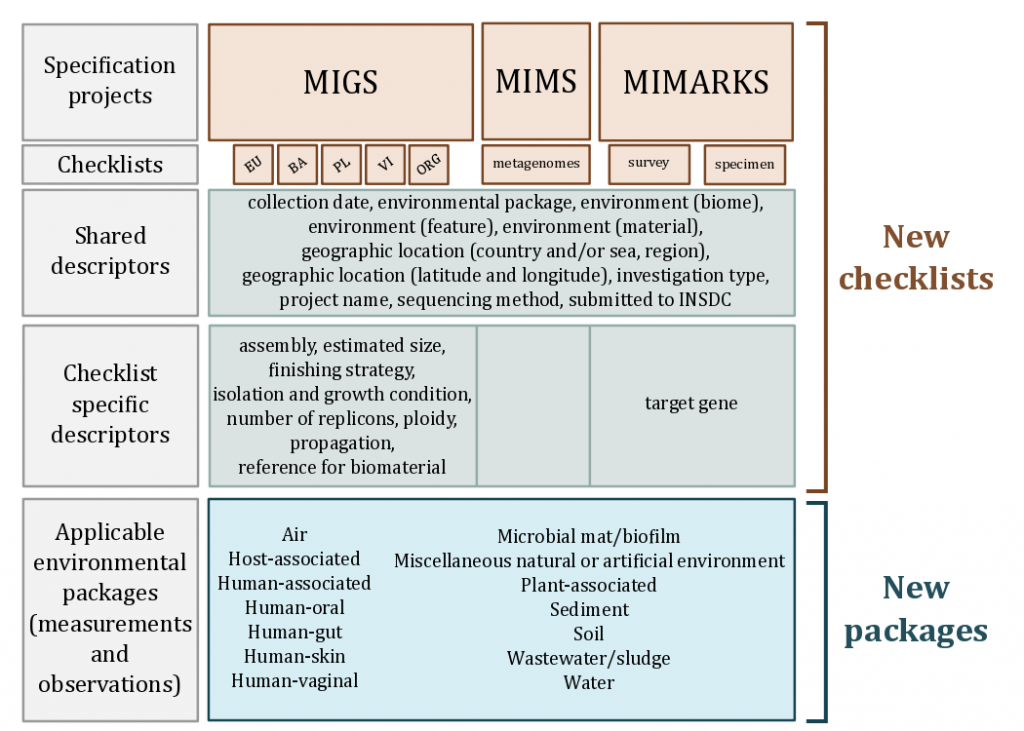
For the complete documentation on each checklist, extension and individual term please visit the MIxS repository content pages.
Current Extensions (packages)
These are the current set of environmental packages:
-
MIxS-agriculture (J. P. Dundore-Arias, et.al 2020 link to paper )
-
MIxS-air (Yilmaz et al. 2011 link to paper )
-
MIxS-built environment (Glass et al. 2014 link to paper )
-
MIxS-host-associated (Yilmaz et al. 2011 link to paper )
-
MIxS-human-associated (Yilmaz et al. 2011 link to paper )
-
MIxS-human-gut (Yilmaz et al. 2011 link to paper )
-
MIxS-human-oral (Yilmaz et al. 2011 link to paper )
-
MIxS-human-skin (Yilmaz et al. 2011 link to paper )
-
MIxS-human-vaginal (Yilmaz et al. 2011 link to paper )
-
MIxS-hydrocarbon resources-cores (Tsesmetzis et al. 2016 link to paper )
-
MIxS-hydrocarbon resources-fluids/swabs (Tsesmetzis et al. 2016 link to paper )
-
MIxS-microbial mat/biofilm (Yilmaz et al. 2011 link to paper )
-
MIxS-miscellaneous natural or artificial environment (Yilmaz et al. 2011 link to paper )
-
MIxS-plant-associated (Yilmaz et.al. 2011 link to paper )
-
MIxS-sediment (Yilmaz et.al. 2011 link to paper )
-
MIxS-soil (Yilmaz et.al. 2011 link to paper )
-
MIxS-symbiont-associated (Jorge et al., 2022 link to paper )
-
MIxS-wastewater/sludge (Yilmaz et.al. 2011 link to paper )
-
MIxS-water (Yilmaz et.al. 2011 link to paper )
Planned Extensions
Four further extensions to MIxS checklists are currently under development:
- Plant genomes, Ramona Walls (rlwalls2008@gmail.com)
- Pathogen package, Lynn Schriml (lschriml@som.umaryland.edu)
- Single cell genomes and genome assemblies from metagenomes, Tanja Woyke (twoyke@lbl.gov) and Susannah Tringe (SGTringe@lbl.gov)
- Urobiome, Lisa Karstens.
- Ancient DNA extension MinAS, James Fellows Yates. Project proposal
Developing Extensions
Proposing and developing new extensions is simple. Development of new extensions should be an open and iterative process engaging the GSC community, the GSC’s MIxS developers, and finally stakeholders across your community.
We can recommend the following steps to anyone interested in developing new extensions:
- identify reported metadata from literature and/or sequence databases
- solicit community input
- formulate an initial checklist or package
- present initial list and rationale behind the extension at a GSC meeting. Alternatively, solicit feedback via our general mailing list or compliance and interoperability mailing list (gensc-cig[at]googlegroups.com)
- coordinate efforts with the MIxS working group to finalize metadata item descriptions, requirements, and other format issues
- procure final approval from GSC MIxS group leader
Once the above steps are completed, the new extension is integrated into our common MIxS database and maintained there. Furthermore, with a new release of MIxS checklists, the extension will also be included.
