Genomic Standards Consortium
The Genomic Standards Consortium (GSC) is an open-membership working body formed in September 2005. The aim of the GSC is making genomic data discoverable. The GSC enables genomic data integration, discovery and comparison through international community-driven standards.
This project is maintained by GenomicsStandardsConsortium
Background
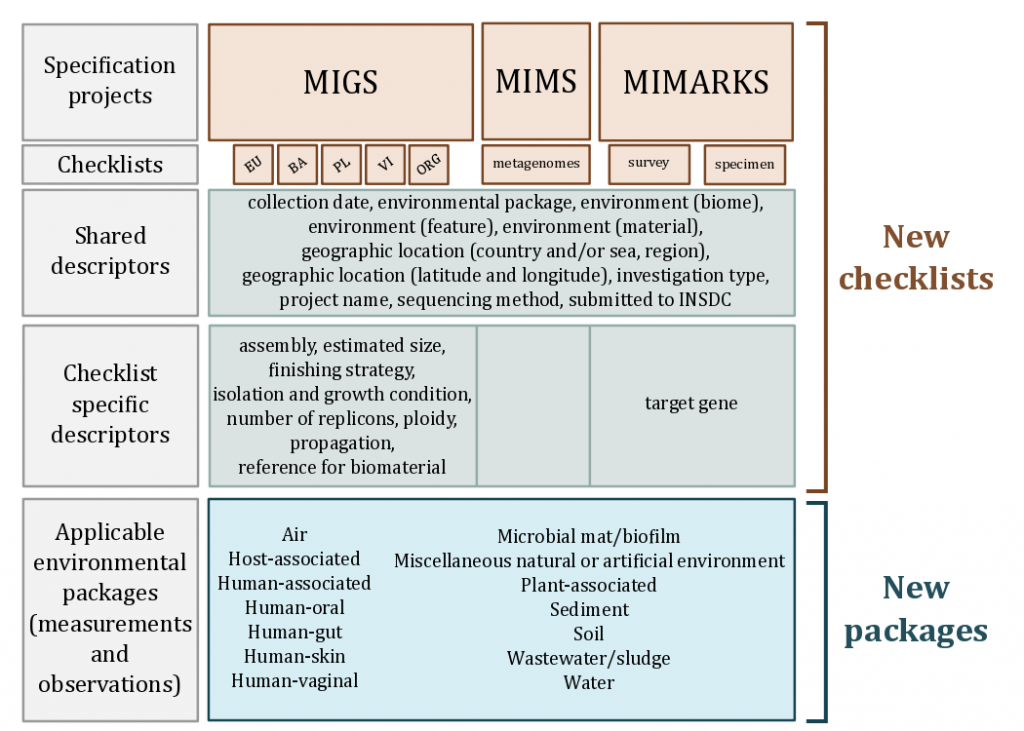
The first MIGS specification enabled description of the complete range of possible genomes (eukaryotes, bacteria, archaea, plasmids, viruses, organelles) and metagenomes. With core descriptors to include information about the origins of the nucleic acid sequence (genome), its environment (latitude and longitude, date and time of sampling and habitat) and sequence processing (sequencing and assembly methods). Since then the GSC have expanded the family of checklists to not only allow a more comprehensive cataloging of metadata per sequence, but also to include sequence types that were not feasible at the time such as single cell sequencing (MISAG) and metagenome assembly sequences (MIMAG).
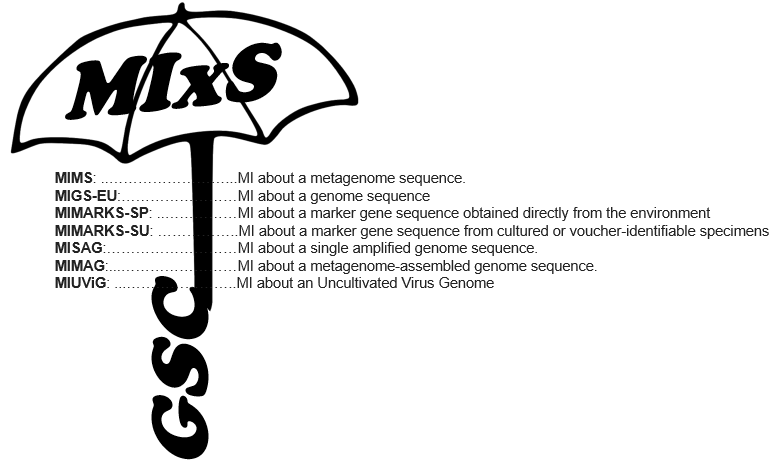
This suite of checklists has become known as “MIxS” (Minimal Information About (X) Any Sequence, pronounced MIX-ess) standards introduced the reporting of a breadth of environment-specific metadata variables to augment the genome-specific checklists. Thus enabling the mix and matching of genome checklists and environmental-specific packages.
The concept of the MIxS suite of standards has subsequently evolved to represent an overarching framework for checklists and environmental packages, creating a single entry point to all minimum information genome checklists from the GSC and to the environmental metadata extensions that together create environmental packages.
MIxS includes the technology-specific checklists from the previous MIGS and MIMS standards, provides a way of introducing additional checklists such as MIMARKS, and also allows annotation of sample data using environmental packages. With this modular and extensible framework, both horizontal (i.e new environmental packages) and vertical extension (i.e. new checklists) development is possible and encouraged.
Minimum Information about any Sequence (MIxS)
The current versions of the GSC family of minimum information standards (MIxS - Minimum Information about any (x) Sequence) are available below. The GSC have provided the information in sections to enable users to understand the relationships between the various levels of complexity, and provided a terminology section (below) to assist comprehension.
In addition, the GSC maintain a GitHub repository where the next versions are being developed and discussed, please feel free to join the discussion using the github issue tracker. The GitHub Wiki in that repository also contains some more techincal specifications.
You can find all previous versions of the checklists in our mixs-legacy GitHub repository.
Terminology (GSC parlance):
Checklist
A checklist is a collection of metadata terms to minimally describe the sampling and sequencing method of a specimen used to generate a nucleotide sequence.
- See the checklist documentation page for more details on individual checklists.
Extension (previously known as Environmental package)
An extension is the collection of recommended metadata terms as developed by community experts, describing the specific context under which a sample was collected.
See the extensions documentation page for more details on available extensions.
Combination (previously known as packages)
A combination of a Checklist with an Extension.
See the combinations documentation page for more details on available combinations.
Term
A term is an individual metadata attribute used to describe any aspect of a sample, sequence, environment or methodology. This is the granular element of any checklist or extension.
Each term is uniquely defined and given its own URI. See the terms page for a complete searchable list of all terms defined within the GSC MIxS checklists and extensions.
Term attributes
Item
The full name of item as it appears in the publication
MIXS ID
The resolvable globally unique persistent identifier associated with a Term, Extension, Checklist or Combination, the unique identifier of an item as a CURIE (compact URI) with the prefix ‘MIXS:’ these expand to https://w3id.org/gensc/mixs/
There are also autogenerated MakeDocs pages for all Checklists, Extensions and Terms that can be found here.
Structured comment name
name of a checklist item as it will appear in GenBank structured comments
Definition
The description of the item, including links to ontologies and other resources that can be used to fill in values for the item
expected value
short description and/or expected value of an item
Value syntax
the proper syntax for writing the value for a given item
Example
Examples of values for an item
Section
The grouping of a metadata term into a specific category. The current sections are;
- **Investigation** ; terms that relate to the project and linking it to other electronic resources.
- **Nucleic acid sequence source** ; terms that are about where and how the DNA sample was created.
- **Sequencing** ; terms that relate to how the DNA sample was processed for sequencing.
- **Environment** ; terms that relate to the environment from which the sample/DNA was obtained.
There are additional documentation about Sections here
Requirement
Information about whether an item is;
- Mandatory (M)
- Conditional mandatory (C)
- Optional (X)
- Environment-dependent (E)
- Not applicable (-)
Preferred units
a unit suggestion if a measurement value is given
Occurrence
indicates whether a given item can be used only once (1), multiple times (m), or none (0)
Core (a deprecated word)
The initial set of GSC metadata standards included a set of ten metadata terms that were required to be included in every checklist at that time, as these terms were considered to be vital for the reporting of metadata for any genome sequence. With the development of additional checklists the concept of ‘core’ evolved to be a list of terms recommended during the development of new checklists, and included terms which were Mandatory or Conditional Mandatory in ANY individual checklist. While this concept proved useful during the developmental phase of checklists before the implementation of the Link-ML model, we feel it is no longer adequately defined and henceforth will no longer be using this phrase.
Compliance and Implementation
Details of how to comply with the standard, and which repositories and institutions are currently making use of them (adopters).
Legacy
You can find all previous versions of the checklists in our mixs-legacy GitHub repository.
