Genomic Standards Consortium
The Genomic Standards Consortium (GSC) is an open-membership working body formed in September 2005. The aim of the GSC is making genomic data discoverable. The GSC enables genomic data integration, discovery and comparison through international community-driven standards.
This project is maintained by GenomicsStandardsConsortium
The ISA framework (ISA-Tab format, ISA software suite and ISA Commons - the community)
Project Lead
Philippe Rocca-Serra, Susanna-Assunta Sansone (University of Oxford e-Research Centre, Oxford, UK)
Initial Team Members
Initial group has co-authored a Nature Genetics publication.
This includes GSC board members (Dawn Field, Jack Gilbert and Linda Amaral-Zettler), others groups also operating in the (meta)genomics domain and beyond such as GigaScience/GigaDB at BGI, the newly launched EBI MetaboLights database and several NIH-funded efforts at Harvard (e.g. the Stem Cell Discovery Engine, published in NAR. Existing and new community using the ISA framework are referred to as the ISA Commons and are listed on the website.
Elevator pitch: A domain agnostic metadata tracking framework that enables collection, curation and management of experiments (with one or several assays), using relevant community standards, in an increasingly diverse set of life science domains.
Project Summary: The open ISA framework belongs to its community of users and contributors, who are assisted by a dedicated team (led by Sansone and Rocca-Serra) that has supported its open developments since 2007.
At the heart of this open source framework is the general-purpose ISA-Tab file format, built on the ‘Investigation’ (the project context), ‘Study’ (a unit of research) and ‘Assay’ (analytical measurement) metadata categories. The extensible, hierarchical structure of this format enables the representation of studies employing one or a combination of technologies, focusing on the description of its experimental metadata (i.e. sample characteristics, technology and measurement types, sample-to-data relationships). The ISA-tab format can be ‘configured’ to follow the requirement of one or more community minimal requirement checklists and one or more terms from ontology (also tracking their provenance). The ISA software suite - the second element of this framework – acts to configure, create and edit ISA-Tab files, store, serve and convert them to a growing number of other related format used by public repositories, including SRAxml. Both format and software suite are described in a Bioinformatics publication; specification, documentation and source code are available from the ISA tools website.
The ISA Commons includes (1) a growing ecosystem of public and internal resources that use the ISA-Tab file format, and/or is powered by one or more component of the ISA software, but also (2) other grass-root standards groups that leverage on the ISA-Tab format, such as the NCI nanotechnology group.
Which existing projects, if any, does this one replace/complement/subsume? The ISA framework is an unique, generic-purpose and configurable format and a set of related tools. By focusing on the description of its experimental metadata (i.e. sample characteristics, technology and measurement types, sample-to-data relationships), the ISA framework is already serving an increasingly diverse set of domains including environmental health, environmental genomics, metabolomics, (meta)genomics, proteomics, stem cell discovery, systems biology, transcriptomics and toxicogenomics, but also communities working to characterize nucleic acid structures and to build a library of cellular signatures.
The ISA framework has been developed via several open workshop held since 2007, including representatives of all existing and relevant community groups, and in synergy with other related, domain-agnostic effort such asFUGE.
How does this project fit into GSC’s mission statement (might also expand it)? ISA is an open source, widely used framework that also enable compliance to MIXs standards.
Will you start a GSC working group? The ISA Commons is an existing, large community, already including GSC members and also The work has always been present at all GSC meetings and it will continue to happen. Also as more groups operating in the (meta)genomics area are joining the ISA Commons, but also because there are groups in GSC that are or will progressively use other assays and the ISA framework will become for them increasingly relevant.
How do you wish to further engage the GSC (recruit members to project, get consultation, link to other GSC projects, etc)? We welcome all interested parties to the ISA Commons.
Do you already have a website or do you wish to create a home page for the project in the GSC website? What other resources might you like from what the GSC can offer? Websites, mailing lists and GitHub projects repositories exists for all material (technical and not), communication and dissemination. But we look forwards to have formal ‘links’ with GSC, as the Board works to finalize the strategy and roles for ‘engaging’ with other related efforts.
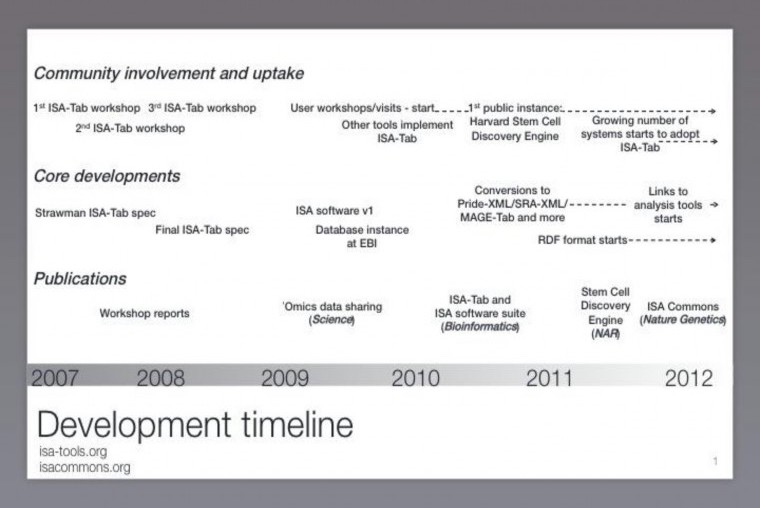
What kind of timeline are you working to for building consensus, releasing first version etc? See development timeline (Appendix 1), including major milestones and current ongoing developments (conversion to RDF and links to analysis tools).
What resources will be required for completion? This is a large project, with a dedicated support team funded mainly by two UK BBSRC resource funds, but with a growing number of communities already contributing time and code extensions, including R&D groups in industry and EU consortia (more on this in a ISA book chapter out in Summer 2012) and groups from public repositories, such as the EBI MetaboLights database. The latter leads an international consortium - the ISA support team is partner in - that has just received funds from the EU also to enhance and extend ISA framework for metabolomics applications.
What are your current plans for publishing/promoting the project?
Technical (Rocca-Serra et al, Bioinformatics, 2010) and community (Sansone et al, Nature Genetics, 2012) papers have been published, and progressively more will come out showing ISA framework in action (first example is the Harvard’s Stem Cell Discovery Engine, Ho Sui et al, NAR Database Issue 2012). Visit to collaborators and new interested parties are ongoing and hands- on workshops will continue.
Have you spoken about the project already within GSC? The work has always been present at all GSC meetings since its onset, making several call for participation and the RCN4GSC has also sponsored visits to GSC members.
References or relevant websites (for further reading)
-
www.isacommons.org (user and contributors’ communities)
-
www.isa-tools.org (specification, code repositories, examples, documentation)
Appendix 1